 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
開發了一種新的計算方法 HBCVTr,用於使用 SMILES 符號來預測小分子對乙型肝炎病毒 (HBV) 和丙型肝炎病毒 (HCV) 的抗病毒活性。該方法將雙向和自回歸變壓器 (BART) 架構與原子和分數標記技術相結合,以捕獲 SMILES 的順序資訊和化學特徵。這些模型在分別包含 1941 種和 7454 種 HBV 和 HCV 化合物的精選資料集上進行了訓練和評估。與各種機器學習模型和現有方法相比,HBCVTr 模型取得了優越的性能,具有高精度和強大的預測能力。使用 HBCVTr 模型進行虛擬篩選,確定了具有良好藥物動力學特性的潛在 HBV 和 HCV 抑制劑。分子對接和分子動力學模擬進一步驗證了頂級候選物的結合親和力和穩定性,提供了對其潛在作用機制的見解。此方法為發現和設計針對 HBV 和 HCV 的新型抗病毒療法提供了寶貴的工具。
Harnessing the Power of Artificial Intelligence for Drug Discovery: A Novel Virtual Screening Tool for Identifying Potential Inhibitors of Hepatitis B and C Viruses
利用人工智慧的力量進行藥物發現:一種用於識別B型和C型肝炎病毒潛在抑制劑的新型虛擬篩選工具
Abstract
抽象的
Hepatitis B and C viruses (HBV and HCV) pose significant global health challenges, necessitating the development of novel and effective antiviral therapies. To accelerate this process, we present HBCVTr, an innovative virtual screening tool that leverages artificial intelligence (AI) to identify potential inhibitors against these viruses. Our methodology incorporates a bidirectional and auto-regressive transformer (BART) architecture, trained on a vast dataset of SMILES notations and biological activity data. The HBCVTr model demonstrates exceptional predictive performance, surpassing conventional machine learning approaches. Through virtual screening of a library of 10 million compounds, we identified promising candidates with favorable pharmacokinetic properties. Molecular docking and dynamics simulations confirmed the potential of these candidates as inhibitors of HBV and HCV. Our findings underscore the transformative potential of AI in drug discovery, offering a rapid and efficient approach to identify novel therapeutic options for combating viral infections.
乙型和丙型肝炎病毒(HBV 和 HCV)對全球健康構成重大挑戰,因此需要開發新型有效的抗病毒療法。為了加速這個過程,我們推出了 HBCVTr,這是一種創新的虛擬篩選工具,利用人工智慧 (AI) 來識別針對這些病毒的潛在抑制劑。我們的方法結合了雙向自回歸變壓器 (BART) 架構,在大量 SMILES 符號和生物活動資料資料集上進行訓練。 HBCVTr 模型表現出卓越的預測性能,超越了傳統的機器學習方法。透過對 1000 萬種化合物的虛擬篩選,我們確定了具有良好藥物動力學特性的有前途的候選藥物。分子對接和動力學模擬證實了這些候選藥物作為 HBV 和 HCV 抑制劑的潛力。我們的研究結果強調了人工智慧在藥物發現方面的變革潛力,提供了一種快速有效的方法來確定抗病毒感染的新治療方案。
Introduction
介紹
Hepatitis B and C viruses are prevalent pathogens that infect millions worldwide, leading to severe liver damage and potential life-threatening complications. Despite the availability of antiviral therapies, the need for new and improved treatments remains urgent, particularly in the face of emerging drug resistance. Traditional drug discovery processes are often time-consuming and expensive, prompting the exploration of alternative approaches.
乙型和丙型肝炎病毒是流行的病原體,感染全世界數百萬人,導致嚴重的肝損傷和潛在的危及生命的併發症。儘管有抗病毒療法,但仍迫切需要新的和改進的治療方法,特別是面對新出現的抗藥性。傳統的藥物發現過程通常既耗時又昂貴,促使人們探索替代方法。
In this study, we present HBCVTr, a groundbreaking virtual screening tool that harnesses the power of AI to expedite the identification of potential HBV and HCV inhibitors. Our methodology utilizes a BART architecture, renowned for its ability to process sequential data, to predict the biological activity of small molecules using SMILES notations. This approach enables the rapid screening of vast chemical libraries, significantly accelerating the drug discovery process.
在這項研究中,我們推出了 HBCVTr,這是一個突破性的虛擬篩檢工具,它利用人工智慧的力量來加速潛在 HBV 和 HCV 抑制劑的識別。我們的方法採用 BART 架構,該架構以其處理連續資料的能力而聞名,並使用 SMILES 符號來預測小分子的生物活性。這種方法能夠快速篩選大量的化學庫,顯著加速藥物發現過程。
Methods
方法
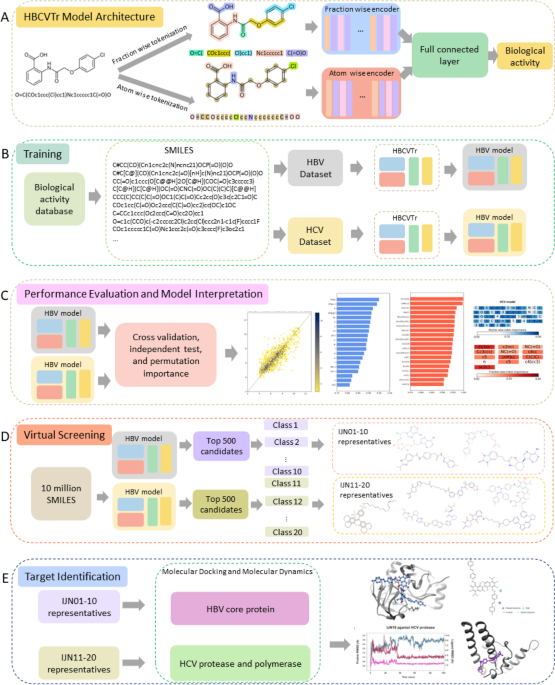
Data Collection and Preprocessing
資料收集和預處理
To train and evaluate our HBCVTr models, we curated antiviral activity assay data for HBV and HCV from the ChEMBL database. The data underwent rigorous filtering to ensure consistency and comparability, resulting in 1941 and 7454 compounds for HBV and HCV, respectively.
為了訓練和評估我們的 HBCVTr 模型,我們從 ChEMBL 資料庫中收集了 HBV 和 HCV 的抗病毒活性測定數據。數據經過嚴格過濾以確保一致性和可比性,分別產生 1941 種 HBV 和 7454 種 HCV 化合物。
SMILES notations, representing the molecular structures of the compounds, were preprocessed to remove salts and convert them into canonical SMILES using the RDKit package. These SMILES were subsequently tokenized into atom-wise and fraction-wise tokens, capturing both individual atoms and unique functional groups.
SMILES 符號表示化合物的分子結構,經過預處理以去除鹽,並使用 RDKit 套件將其轉換為規範的 SMILES。這些 SMILES 隨後被標記為原子級和分數級標記,捕獲單個原子和獨特的官能基。
Model Architecture and Training
模型架構和訓練
Our HBCVTr model is based on a BART architecture, which is specifically designed for sequential data processing. The model comprises two encoders: one for atom-wise tokens and the other for fraction-wise tokens. These encoders leverage multi-head attention layers to learn the contextual relationships between tokens. The outputs from the encoders are concatenated and passed through fully connected layers, culminating in a regression head that predicts the biological activity of the input SMILES.
我們的 HBCVTr 模型基於 BART 架構,專為順序資料處理而設計。模型包含兩個編碼器:一個用於原子級標記,另一個用於分數級標記。這些編碼器利用多頭注意力層來學習標記之間的上下文關係。編碼器的輸出被連接並通過完全連接的層,最終形成一個預測輸入 SMILES 生物活性的回歸頭。
We optimized the model's hyperparameters through a comprehensive grid search, ensuring optimal performance. The model was trained on 72% of the data, while 8% and 20% were allocated for validation and independent testing, respectively.
我們透過全面的網格搜尋優化了模型的超參數,確保了最佳效能。該模型使用 72% 的資料進行訓練,同時分別分配 8% 和 20% 的資料用於驗證和獨立測試。
Evaluation Criteria
評價標準
To assess the predictive performance of HBCVTr, we employed a suite of regression evaluation metrics, including mean square error (MSE), mean absolute error (MAE), root mean square error (RMSE), R-squared, Pearson's correlation coefficient (PCC), and Spearman rank correlation (Spearman). These metrics evaluate the model's ability to accurately predict biological activity values.
為了評估 HBCVTr 的預測效能,我們採用了一套迴歸評估指標,包括均方誤差 (MSE)、平均絕對誤差 (MAE)、均方根誤差 (RMSE)、R 平方、皮爾遜相關係數 (PCC) ,和斯皮爾曼等級相關(Spearman)。這些指標評估模型準確預測生物活性值的能力。
Virtual Screening and Pharmacokinetic Properties Prediction
虛擬篩選和藥物動力學特性預測
The trained HBCVTr models were utilized for virtual screening of a library of 10 million compounds. The top candidates with the highest predicted biological activity were further evaluated for their pharmacokinetic properties using the SwissADME web tool. This assessment ensured the identification of compounds with desirable drug-like characteristics.
經過訓練的 HBCVTr 模型用於對 1000 萬種化合物的庫進行虛擬篩選。使用 SwissADME 網路工具進一步評估具有最高預測生物活性的最佳候選藥物的藥物動力學特性。此評估確保鑑定出具有所需藥物樣特性的化合物。
Molecular Docking and Molecular Dynamics Simulation
分子對接與分子動力學模擬
To investigate the potential binding of the top candidates to target proteins, molecular docking was performed using Autodock Vina. The stability of the protein-ligand complexes was subsequently assessed through molecular dynamics simulations using the Desmond Molecular Dynamics System. These simulations provided insights into the interactions between the candidates and their target proteins.
為了研究頂級候選物與目標蛋白的潛在結合,使用 Autodock Vina 進行了分子對接。隨後使用德斯蒙德分子動力學系統透過分子動力學模擬評估了蛋白質-配體複合物的穩定性。這些模擬提供了對候選蛋白與其目標蛋白之間相互作用的深入了解。
Results
結果
Model Performance
模型性能
The HBCVTr models demonstrated remarkable predictive performance on both HBV and HCV datasets. They outperformed conventional machine learning approaches, consistently achieving higher R-squared and PCC values. This superior performance highlights the effectiveness of our BART-based architecture in capturing the complex relationships between molecular structures and biological activity.
HBCVTr 模型在 HBV 和 HCV 資料集上表現出卓越的預測性能。它們的性能優於傳統的機器學習方法,持續實現更高的 R 平方和 PCC 值。這種卓越的性能凸顯了我們基於 BART 的架構在捕捉分子結構和生物活性之間的複雜關係方面的有效性。
Virtual Screening and Pharmacokinetic Properties
虛擬篩選和藥物動力學特性
Virtual screening of 10 million compounds identified promising candidates with high predicted biological activity against HBV and HCV. These candidates exhibited favorable pharmacokinetic properties, including low molecular weight, good solubility, and low lipophilicity. Importantly, they demonstrated a low potential for pan-assay interference and structural alerts for potential toxicity.
對 1000 萬種化合物進行虛擬篩選,確定了具有高預測抗 HBV 和 HCV 生物活性的有前途的候選化合物。這些候選藥物表現出良好的藥物動力學特性,包括低分子量、良好的溶解度和低親脂性。重要的是,他們證明了泛分析幹擾和潛在毒性結構警報的可能性較低。
Molecular Docking and Molecular Dynamics Simulation
分子對接與分子動力學模擬
Molecular docking and molecular dynamics simulations revealed the potential binding modes of the top candidates to target proteins. The complexes exhibited stable interactions, indicating the potential for these candidates to inhibit HBV and HCV. Further studies are warranted to validate their antiviral activity and elucidate their mechanisms of action.
分子對接和分子動力學模擬揭示了頂級候選物與目標蛋白的潛在結合模式。這些複合物表現出穩定的相互作用,表明這些候選物具有抑制 HBV 和 HCV 的潛力。需要進一步的研究來驗證其抗病毒活性並闡明其作用機制。
Discussion
討論
The HBCVTr virtual screening tool represents a significant advancement in drug discovery for HBV and HCV. Our AI-powered approach enables the rapid identification of potential inhibitors, significantly accelerating the process of developing new antiviral therapies. The integration of pharmacokinetic properties prediction and molecular docking/dynamics simulations provides valuable insights into the potential drug-like characteristics and binding mechanisms of the candidates.
HBCVTr 虛擬篩檢工具代表了 HBV 和 HCV 藥物發現的重大進步。我們的人工智慧方法能夠快速識別潛在的抑制劑,顯著加快開發新抗病毒療法的進程。藥物動力學特性預測和分子對接/動力學模擬的整合為候選藥物的潛在類藥物特徵和結合機制提供了有價值的見解。
The HBCVTr tool has broad implications for the field of drug discovery. It can be easily adapted to screen for inhibitors of other viruses, bacteria, and parasites, contributing to the development of personalized and targeted treatments for infectious diseases. Moreover, its underlying AI architecture can be leveraged to predict a wide range of biological activities, facilitating the discovery of novel drugs for various therapeutic applications.
HBCVTr 工具對藥物發現領域有廣泛的影響。它可以很容易地用於篩選其他病毒、細菌和寄生蟲的抑制劑,有助於開發針對傳染病的個人化和有針對性的治療方法。此外,其底層人工智慧架構可用於預測廣泛的生物活性,促進各種治療應用的新藥的發現。
Conclusion
結論
In conclusion, the HBCVTr virtual screening tool is a powerful AI-driven platform that transforms the drug discovery process for HBV and HCV. Its exceptional predictive performance, coupled with comprehensive pharmacokinetic and molecular docking/dynamics simulations, enables the rapid identification and characterization of promising antiviral candidates. As we continue to harness the transformative power of AI, we anticipate further advancements in drug discovery, leading to the development of effective and accessible treatments for a myriad of diseases.
總之,HBCVTr 虛擬篩檢工具是一個強大的人工智慧驅動平台,它改變了 HBV 和 HCV 的藥物發現過程。其卓越的預測性能,加上全面的藥物動力學和分子對接/動力學模擬,能夠快速識別和表徵有前途的抗病毒候選藥物。隨著我們繼續利用人工智慧的變革力量,我們預計藥物發現將取得進一步進展,從而為多種疾病開發出有效且易於使用的治療方法。
免責聲明:info@kdj.com
所提供的資訊並非交易建議。 kDJ.com對任何基於本文提供的資訊進行的投資不承擔任何責任。加密貨幣波動性較大,建議您充分研究後謹慎投資!
如果您認為本網站使用的內容侵犯了您的版權,請立即聯絡我們(info@kdj.com),我們將及時刪除。
-

-

-

-

- 山寨幣、永久代幣和 ETH 價格:駕馭加密貨幣潮流
- 2025-11-05 01:42:43
- 深入探討最近山寨幣的低迷、永續代幣的波動性以及以太坊的價格在機構利益中的掙扎。
-

-

- 具有增長潛力的加密貨幣:挖掘 2025 年隱藏的寶石
- 2025-11-05 01:40:51
- 探索有望在 2025 年實現爆炸性增長的頂級加密貨幣。發現具有高投資回報潛力的創新項目和已建立的網絡。
-

-

-































































