|
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
Cryptocurrency News Articles
Harnessing AI to Combat Hepatitis: Virtual Screening Tool Identifies Dormant Inhibitors
Apr 23, 2024 at 05:02 am
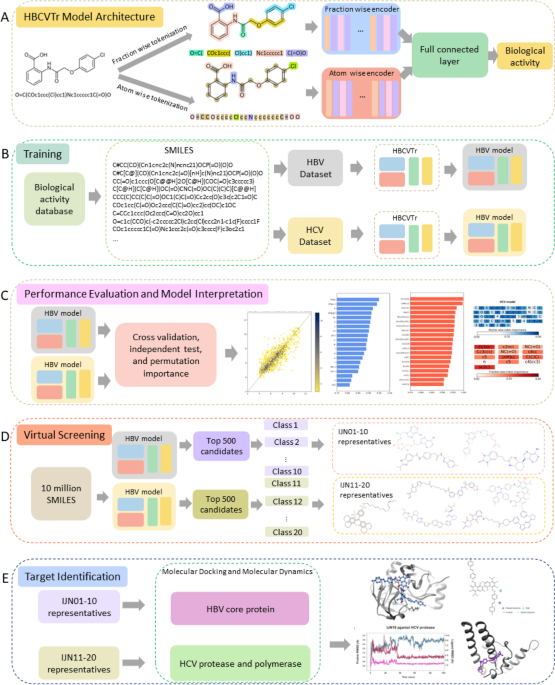
A new computational methodology, HBCVTr, was developed to predict the antiviral activity of small molecules against hepatitis B virus (HBV) and hepatitis C virus (HCV) using their SMILES notations. This methodology combines bidirectional and auto-regressive transformer (BART) architecture with atom-wise and fraction-wise tokenization techniques to capture the sequential information and chemical features of SMILES. The models were trained and evaluated on curated datasets containing 1941 and 7454 compounds for HBV and HCV, respectively. The HBCVTr models achieved superior performance compared to various machine learning models and existing methods, with high accuracy and robust prediction capabilities. Virtual screening using the HBCVTr models identified potential HBV and HCV inhibitors with promising pharmacokinetic properties. Molecular docking and molecular dynamics simulations further validated the binding affinity and stability of the top candidates, providing insights into their potential mechanisms of action. This methodology offers a valuable tool for discovering and designing novel antiviral therapies against HBV and HCV.
Harnessing the Power of Artificial Intelligence for Drug Discovery: A Novel Virtual Screening Tool for Identifying Potential Inhibitors of Hepatitis B and C Viruses
Abstract
Hepatitis B and C viruses (HBV and HCV) pose significant global health challenges, necessitating the development of novel and effective antiviral therapies. To accelerate this process, we present HBCVTr, an innovative virtual screening tool that leverages artificial intelligence (AI) to identify potential inhibitors against these viruses. Our methodology incorporates a bidirectional and auto-regressive transformer (BART) architecture, trained on a vast dataset of SMILES notations and biological activity data. The HBCVTr model demonstrates exceptional predictive performance, surpassing conventional machine learning approaches. Through virtual screening of a library of 10 million compounds, we identified promising candidates with favorable pharmacokinetic properties. Molecular docking and dynamics simulations confirmed the potential of these candidates as inhibitors of HBV and HCV. Our findings underscore the transformative potential of AI in drug discovery, offering a rapid and efficient approach to identify novel therapeutic options for combating viral infections.
Introduction
Hepatitis B and C viruses are prevalent pathogens that infect millions worldwide, leading to severe liver damage and potential life-threatening complications. Despite the availability of antiviral therapies, the need for new and improved treatments remains urgent, particularly in the face of emerging drug resistance. Traditional drug discovery processes are often time-consuming and expensive, prompting the exploration of alternative approaches.
In this study, we present HBCVTr, a groundbreaking virtual screening tool that harnesses the power of AI to expedite the identification of potential HBV and HCV inhibitors. Our methodology utilizes a BART architecture, renowned for its ability to process sequential data, to predict the biological activity of small molecules using SMILES notations. This approach enables the rapid screening of vast chemical libraries, significantly accelerating the drug discovery process.
Methods
Data Collection and Preprocessing
To train and evaluate our HBCVTr models, we curated antiviral activity assay data for HBV and HCV from the ChEMBL database. The data underwent rigorous filtering to ensure consistency and comparability, resulting in 1941 and 7454 compounds for HBV and HCV, respectively.
SMILES notations, representing the molecular structures of the compounds, were preprocessed to remove salts and convert them into canonical SMILES using the RDKit package. These SMILES were subsequently tokenized into atom-wise and fraction-wise tokens, capturing both individual atoms and unique functional groups.
Model Architecture and Training
Our HBCVTr model is based on a BART architecture, which is specifically designed for sequential data processing. The model comprises two encoders: one for atom-wise tokens and the other for fraction-wise tokens. These encoders leverage multi-head attention layers to learn the contextual relationships between tokens. The outputs from the encoders are concatenated and passed through fully connected layers, culminating in a regression head that predicts the biological activity of the input SMILES.
We optimized the model's hyperparameters through a comprehensive grid search, ensuring optimal performance. The model was trained on 72% of the data, while 8% and 20% were allocated for validation and independent testing, respectively.
Evaluation Criteria
To assess the predictive performance of HBCVTr, we employed a suite of regression evaluation metrics, including mean square error (MSE), mean absolute error (MAE), root mean square error (RMSE), R-squared, Pearson's correlation coefficient (PCC), and Spearman rank correlation (Spearman). These metrics evaluate the model's ability to accurately predict biological activity values.
Virtual Screening and Pharmacokinetic Properties Prediction
The trained HBCVTr models were utilized for virtual screening of a library of 10 million compounds. The top candidates with the highest predicted biological activity were further evaluated for their pharmacokinetic properties using the SwissADME web tool. This assessment ensured the identification of compounds with desirable drug-like characteristics.
Molecular Docking and Molecular Dynamics Simulation
To investigate the potential binding of the top candidates to target proteins, molecular docking was performed using Autodock Vina. The stability of the protein-ligand complexes was subsequently assessed through molecular dynamics simulations using the Desmond Molecular Dynamics System. These simulations provided insights into the interactions between the candidates and their target proteins.
Results
Model Performance
The HBCVTr models demonstrated remarkable predictive performance on both HBV and HCV datasets. They outperformed conventional machine learning approaches, consistently achieving higher R-squared and PCC values. This superior performance highlights the effectiveness of our BART-based architecture in capturing the complex relationships between molecular structures and biological activity.
Virtual Screening and Pharmacokinetic Properties
Virtual screening of 10 million compounds identified promising candidates with high predicted biological activity against HBV and HCV. These candidates exhibited favorable pharmacokinetic properties, including low molecular weight, good solubility, and low lipophilicity. Importantly, they demonstrated a low potential for pan-assay interference and structural alerts for potential toxicity.
Molecular Docking and Molecular Dynamics Simulation
Molecular docking and molecular dynamics simulations revealed the potential binding modes of the top candidates to target proteins. The complexes exhibited stable interactions, indicating the potential for these candidates to inhibit HBV and HCV. Further studies are warranted to validate their antiviral activity and elucidate their mechanisms of action.
Discussion
The HBCVTr virtual screening tool represents a significant advancement in drug discovery for HBV and HCV. Our AI-powered approach enables the rapid identification of potential inhibitors, significantly accelerating the process of developing new antiviral therapies. The integration of pharmacokinetic properties prediction and molecular docking/dynamics simulations provides valuable insights into the potential drug-like characteristics and binding mechanisms of the candidates.
The HBCVTr tool has broad implications for the field of drug discovery. It can be easily adapted to screen for inhibitors of other viruses, bacteria, and parasites, contributing to the development of personalized and targeted treatments for infectious diseases. Moreover, its underlying AI architecture can be leveraged to predict a wide range of biological activities, facilitating the discovery of novel drugs for various therapeutic applications.
Conclusion
In conclusion, the HBCVTr virtual screening tool is a powerful AI-driven platform that transforms the drug discovery process for HBV and HCV. Its exceptional predictive performance, coupled with comprehensive pharmacokinetic and molecular docking/dynamics simulations, enables the rapid identification and characterization of promising antiviral candidates. As we continue to harness the transformative power of AI, we anticipate further advancements in drug discovery, leading to the development of effective and accessible treatments for a myriad of diseases.
Disclaimer:info@kdj.com
The information provided is not trading advice. kdj.com does not assume any responsibility for any investments made based on the information provided in this article. Cryptocurrencies are highly volatile and it is highly recommended that you invest with caution after thorough research!
If you believe that the content used on this website infringes your copyright, please contact us immediately (info@kdj.com) and we will delete it promptly.
-

-

-

-

-

- RWA Tokenization Platform Coldware (COLD) Surges 80% as Sui (SUI) and Avalanche (AVAX) Struggle
- Apr 04, 2025 at 03:10 am
- In the ever-evolving world of cryptocurrency, some tokens rise to prominence while others experience major setbacks. Recent developments have seen Sui (SUI) fall by 4.43%, and Avalanche (AVAX) struggle with a notable 13.83% drop.
-

-

-

-

- Trump's New Global Tariff Regime Reshapes Investor Sentiment, Triggering Sharp Corrections Across Risk Assets
- Apr 04, 2025 at 03:00 am
- Market analysts say President Donald Trump's newly announced global tariff regime is already reshaping investor sentiment, triggering sharp corrections across risk assets.